|
IPIM
|
Module containing the objects and methods to process chemical reactions. More...
Public Member Functions | |
| subroutine | read_reactions_file () |
| Interface to initialize spec type from element type (species). Then call reac_readreac routine and create reac type called reactions. More... | |
| subroutine | reac_readreac (wkdir, reacfile, sp, re) |
| Read the input reactions.dat file and parse each reaction. More... | |
| subroutine | reac_reacinfo (re) |
| Print information on the reactions. More... | |
| subroutine | reac_testreac (sp, re) |
| Tests if the reaction is kept or not by comparing the reactants and the species list. More... | |
| subroutine | old_reac_rrate (rtype, ncoeff, coeff, T, P, T_o, N_o, k_r) |
| Calculates the reaction rate for a given reaction type. More... | |
| subroutine | old_reac_getthirdspec (sp, ptr, nnr, nr, nnu, nu, nc, idx) |
| Gives the concentration of the third reactant in a 3-species reaction. More... | |
| subroutine | reac_fillmatrix (sp, re, nr, nu, T, P) |
| Fills the reaction matrix Mij and the reaction explicit vector Si. More... | |
| subroutine | reac_photodiss (sp, re, flux) |
| subroutine | reac_printinfo (re, unb) |
Module containing the objects and methods to process chemical reactions.
Defines the types and the data structure containing the list of the reactions and their properties. Reactions module.
| subroutine m_reac::old_reac_getthirdspec | ( | type(spec) | sp, |
| type(one_reaction), pointer | ptr, | ||
| nnr, | |||
| nr, | |||
| nnu, | |||
| nu, | |||
| nc, | |||
| idx | |||
| ) |
Gives the concentration of the third reactant in a 3-species reaction.
| sp | Pointer to the current species list |
| ptr | Pointer to the current reaction |
| ptr | Number of reactants in the reaction |
| ptr | Number of species resolved in the code |
| ptr | Number of species non-resolved (given by a model) |
| ptr | Concentations of the resolved species |
| ptr | Concentration of the non-resolved species |
| ptr | Ouput value [ 1:No 3rd reactant | nc:concentration of the 3rd reactant ] |
| ptr | Index of the 3rd reactant |

| subroutine m_reac::old_reac_rrate | ( | rtype, | |
| ncoeff, | |||
| coeff, | |||
| T, | |||
| P, | |||
| real | T_o, | ||
| real | N_o, | ||
| k_r | |||
| ) |
Calculates the reaction rate for a given reaction type.
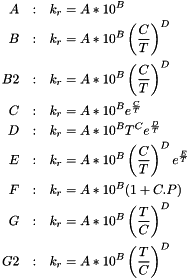
| n_o | Reaction rate |
| n_o | Auxiliar variable |

| subroutine m_reac::reac_fillmatrix | ( | type(spec) | sp, |
| type(reac) | re, | ||
| nr, | |||
| nu, | |||
| T, | |||
| P | |||
| ) |
Fills the reaction matrix Mij and the reaction explicit vector Si.
| re | Local temperature |
| re | Local pressure |
| re | Concentrations of the resolved species |
| re | Concentrations of the non resolved species |
| subroutine m_reac::reac_reacinfo | ( | type(reac) | re | ) |
Print information on the reactions.
| subroutine m_reac::reac_readreac | ( | wkdir, | |
| reacfile, | |||
| type(spec) | sp, | ||
| type(reac) | re | ||
| ) |
Read the input reactions.dat file and parse each reaction.

| subroutine m_reac::reac_testreac | ( | type(spec) | sp, |
| type(reac) | re | ||
| ) |
Tests if the reaction is kept or not by comparing the reactants and the species list.

| subroutine m_reac::read_reactions_file | ( | ) |
Interface to initialize spec type from element type (species). Then call reac_readreac routine and create reac type called reactions.

 1.8.5
1.8.5